Molecular Survey on Symbiodinium of Some Scleractinean Coral Spp. and a Fire Coral sp. along the Red Sea of Egypt
Abstract
The present study introduce an overview on the cladal structure of Symbiodinium population associated with some species of scleractinean corals and fire coral in the Egyptian Red Sea coast and discuss the possible consequences of recent climate changes on coral reefs. Cladal structure of Symbiodinium populations associated with eight keystone species of scleractinean corals and one species of fire coral that collected along Egyptian Red Sea coast, during 2012-2013, had been resolved based on 18S nrDNA and ITS2 genetic markers. Only Symbiodinium subclades C1 and A1 were identified from all examined species. Symbiodinium C1 was the dominant subclade that associated with 61% of coral samples. Results revealed that the studied pocilloporid corals were associated with Symbiodinium C1 and/or A1 while acroporids were only associated with Symbiodinium C1. The present data also indicated that Symbiodinium C1 occurred at high densities than A1 or A1+C1 combination. Because of the relative thermal susceptibility of clades C and A, the current study addresses that the recent climate changes may derive dramatic changes on community structure of coral reefs at the Red Sea.
Author Contributions
Academic Editor: Cesar Amaral, DNA Diagnostic Laboratory, University of the State of Rio de Janeiro, Brazil.
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2019 Muhammad Y.Dosoky, et al.
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
Corals are known to be the typical hosts for symbiotic dinoflagellates of the genus Symbiodinium which are functionally diverse 1. This diversity reflects the functional ecology of different Symbiodinium types that establish different coral-symbiont partnerships in habitats of different conditions. Accordingly, genetic diversity of endosymbionts associated with corals is considered critical for coral life. Many studies had revealed that Symbiodinium clades have different thermal tolerance in which Symbiodinium D is the most thermotolerant and C is the most thermosensitive 2. Though, under environmental stressors, members within the same clade do not necessarily acquire similar physiological responses 3. At finer levels, Symbiodinium types within each clade possess differential secondary thermal tolerances 1, 4. As a consequence, bleaching events are severe at reefs dominated by thermosensitive types 5, 6, 7. Yet, determining the definite thermal tolerance that outlines the susceptibility of corals to bleaching, of many types is still in its infancy 3. However, inspecting genetic composition of coral’s endosymbiotic systems is considered crucial for understanding susceptibility of corals to environmental stressors and predicting the consequences of increasing temperature on coral reefs.
Red Sea coral reefs represent the upper northern limits for coral reefs distribution. Although Red Sea coral reefs are considered unique reefs, they had received little concern about their endosymbiotic systems 8. Only few studies on the genetic diversity of Symbiodinium at the northern tip of the Gulf of Aqaba and at the eastern coasts had been published 9, 10, 11. Despite that the Egyptian coast of the Red Sea extends approximately 1800 km 12, genetic structure of endosymbiotic systems along this western part had not resolved yet, especially in scleractinian corals which are considered the main reef builders. Here, we assess Symbiodinium diversity on the reef building corals host species and one species of fire coral that exist along Egyptian coast of the Red Sea. Coral samples were collected from shallow (1-5) and mid-depth (5-10) habitats. We address geographical differences between coral populations of western Egyptian coast and eastern Saudi Arabian coast of Red Sea in terms of phylogenetic diversity of their dinoflagellate symbionts.
Materials and Methods
Study Sites and Sample Collection
Six sampled locations were chosen along the western Red Sea coast off Egypt. Three locations were tropical in the proper Red Sea (Marsa Samadai: 25° 00′ 50″ N, 34° 55′ 37″ E; Zabargad: 23° 35′ 51″ N, 36° 12′ 31″ E; Rocky Islands: 23° 33′ 47″ N, 36° 14′ 33″ E), or subtropical-temperate in the northern Res Sea (Fanous Reef: 27° 15′ 57″ N, 33° 53′ 30″ E) and Gulf of Aqaba (Marsa Ghozlani: 27° 49′ 20″ N, 34° 15′ 58″ E; Solomon reef: 28° 29′ 10″ N, 34° 30′ 52″ E) see Figure 1 (Table 1). Eight species of scleractinian corals (Acropora digitifera, Acropora humilis, Acropora pharaonis, Pocilloporaverrucosa, Stylophorapistillata,Stylophorawellsi, Porites sp., and Favia sp.), and one species of hydrozoan fire coral (Milleporadichotoma) were selected for the present study. Fragments of mixed samples of coral skeleton and resident symbiont communities were collected by SCUBA during 2012-2013.
Table 1. Symbiodinium types isolated from different coral species along with corresponding NCBI accession numbers.| Host | Site | Symbiodinium Subclade | Accession number |
| A. digitifera | Marsa Ghozlani | C1 | KM066980 |
| A. digitifera | Marsa Samadai | C1 | KM066985 |
| A. humilis | Solomon Reef | C1 | KM066982 |
| A. humilis | Marsa Ghozlani | C1 | KM066981 |
| A. humilis | Fanous Reef | C1 | KM066984 |
| A. humilis | Zabargad Island | C1 | KM066987 |
| A. pharaonis | Solomon Reef | C1 | KM066983 |
| A. pharaonis | Marsa Ghozlani | C1 | KM066979 |
| A. pharaonis | Marsa Samadai | C1 | KM066986 |
| Favia sp. | Marsa Ghozlani | C1 | KM066978 |
| P. verrucosa | Marsa Samadai | A1 | KM066991 |
| S. pistillata | Marsa Ghozlani | A1 | KM066988 |
| S. pistillata | Marsa Samadai | A1 | KM066990 |
| S. wellsi | Marsa Ghozlani | A1 | KM066989 |
Figure 1.Sampling sites along the Egyptian Red Sea.
Samples were collected using a hammer and hollow steel punch (<1 cm2 diameter) penetrating the coral skeleton deeply enough to capture all tissue layers (~3 to 6 mm deep). To minimize the likelihood of repeatedly sampling corals that were the same genetic individual, only conspecifics separated by >10 m were sampled. Samples were taken only from the tops of healthy colonies and identified according to a key reference 13. Immediately, collected tissue was preserved for DNA analysis in saline DMSO buffer 20% (v/v) DMSO 250 mM EDTA saturated NaCl14.
Symbiodinium Phylotyping
DNA of Symbiodinium was extracted using phenol/chloroform methodology 15. Accordingly, 600 µL of 2x CTAB extraction buffer 1.4M NaCl 0.02M EDTA (pH=7.8) 0.1M Tris were added to Symbiodinium pellets. After homogenization, 10 µL of proteinase K (20 mg/ml) were immediately added. Well shacked suspension was then incubated at 65°C for 2 hours with frequent shacking. After that, 600 µL of chloroform isoamyl alcohol 24:1 (v/v) were added twice with an intermediate addition of phenol chloroform isoamyl alcohol 25:24:1 (v/v) to DNA extract (1:1). Prior to each extraction step, solution was shaken for 5 min then centrifuged for 15 min at 13,000 rpm.
DNA was precipitated by adding 1ml of 95% cold ethyl alcohol to the aqueous layer and incubated overnight at -20°C. Samples were then centrifuged for 30 min at 13,000 rpm. DNA pellets were washed twice with 500 µL of 70% cold ethyl alcohol and then centrifuged at 13,000 rpm for 5 min. Pellets of DNA were dried at room temperature and finally dissolved in 100 µL double distilled sterilized water.
The two genetic markers, 18S nrDNA and ITS2, were amplified. The first one, 18S nrDNA, was used to identify Symbiodinium clades in all collected samples (n=98), while ITS2 of a randomly selected subsamples (n=14) was used to identify Symbiodinium subclades.
Each reaction was composed of 1x PCR buffer, 1 mM dNTPs, 3.5 mM MgCl2, 0.2 µM of each forward and reverse primer (18S nrDNA: ss5 and ss3z 16; ITS2: ITS2intfor and ITS2clamp but without GC clamp 17), 0.05U/µL Taq polymerase, 30 ng/µL DNA template, and PCR grade water to final volume of 25µL for 18S nrDNA or 50µL for ITS2. The two markers were amplified under the following conditions: hot start at 94°C for 2min; followed by 35 cycles of 94°C for 1min, 54°C for 1min, and 72°C for 2min; final extension at 72°C for 8min. For Symbiodinium phylotyping, however, 18S nrDNAs were digested by Taq I restriction enzyme. The reaction was composed of 1x PCR buffer, 0.2U/µL Taq I restriction enzyme and 5µL PCR product in a final volume of 25µL with PCR grade water. Isothermal restriction enzyme digestion was performed in thermocycler at 65°C for 30 min. RFLP digests were run on 1.5% agarose gel along with 3kb Mid-Range DNA ladder Jena Bioscience (#M. Gel bands produced by RFLP analysis were analyzed by Gel-Pro Analyzer software v4.0 to estimate molecular size of each band. RFLP profiles in the current study were compared with profiles previously published on Symbiodinium clades using the same primers set and restriction enzyme.
To confirm RFLP analysis results and to identify Symbiodinium subclades upon which phylogenetic relationships would be constructed, ITS2 was sequenced. Part of successfully amplified samples (n= 5) were initially sequenced in the forward and reverse directions by Macrogen (Macrogen Ltd., Korea) while the remainder samples (n= 9) were sequenced by Biotechnology Research Center (Suez Canal University) at forward direction only.
Phylogenetic Analysis
After editing, ITS2 sequences were blasted using MEGABLAST on NCBI GenBank under default algorithm parameters. All the edited ITS2 sequences had been submitted and archived in NCBI GenBank (KM066978.1 ‒ KM066991.1).
ITS2 sequences in the current study and that downloaded from NCBI GenBank were aligned together using ClustalW. Phylogenetic relationships between 55 sequences, among which 14 were resolved by the current study, of Symbiodinium harbored by different taxa of invertebrates along the Red Sea were inferred using p-distance Neighbor-Joining method for 1000 bootstrap replication value 18. Gaps were treated by pairwise deletion.Protodinium simplex, Pelagodiniumbeii, and Polarella glacialis (JN558105, JN558107, and JN558108, respectively) were used for tree rooting. All phylogenetic analyses were conducted using MEGA v6.0 19.
One way ANOVA was performed upon original or transformed data. Data of abnormal distribution, even after transformation, were subjected to Kruskal-Wallis test.
Data Accessibility
All ITS2 sequences in this study were archived at the NCBI GenBank with accession numbers KM066978.1-KM066991.1.
Results
Based on 18S nrDNAs, three different banding patterns were produced by Symbiodinium harbored studied corals. The first banding pattern was characterized by two bands of 1173 and 800 pbs, while the second pattern consisted of three bands of 800, 678, and 288 pbs Figure 2 The third was consisted of a combination of the first and second banding patterns Figure 3 The first and second banding patterns are corresponding to previously published RFLP profiles for Symbiodinium clades C and A, respectively, while the third one is a combination of clades A and C (A+C).
Figure 2.RFLP profiles of 18S nrDNAs digested with Taq I. Clade C profiles (lanes 1-4) was identified from Acropora digitifera , Acropora humilis, Acropora pharaonis, and Stylophora pistillata, while clade A (lane 5) had been identified from Pocillopora verrucosa.
Figure 3.RFLP profiles of 18S nrDNAs digested with Taq I. Clade A isolated from Stylophora pistillata (lanes 1 and 2), Stylophora wellsi (lane 3), Pocillopora verrucosa (lane 4), and Millepora dichotoma (lane 8) while clade C had been isolated from Acropora pharaonis and Acropora humilis (lanes 5 and 6, respectively). A+C combination (lane 7) was isolated from Pocillopora verrucosa.
MEGABLAST search on NCBI GenBank for ITS2 sequences of all Symbiodinium isolated from different coral species confirmed that they relate either to clade A or C. Further sequence data showed that each Symbiodinium clade represented by only one subclade (Figure 4) For clade A, all ITS2 sequences were highly similar to Symbiodinium A1 type (Tridacna maxima, Egypt, Red Sea, GU069005) with a maximum identity of 99%, E value of 3e-120, and query cover of 100%. On the other hand, for clade C, 10 sequences were highly similar to Symbiodinium C1 type (Polycyathus sp., Taiwan, JQ180021) with a maximum identity of 99%, E value of 2e-136, and query cover of 100%. Sequences of isolated Symbiodinium from studied corals were archived in the NCBI GenBank with accession numbers showed in Table 1.
Figure 4.ITS2 electropherogram of Symbiodinium A1 and C1 isolated from Stylophora pistillata and Acropora pharaonis, respectively.
Of nine examined species (n=98), the current genetic survey revealed that 61% of coral samples hosted Symbiodinium C, 33% associated with Symbiodinium A, and only 6% formed symbiosis with A+C combination. Symbiodinium C1 was recorded in acroporid, poritid, faviid, and pocilloporid species, while clade A1 was only detected in the three studied pocilloporid species as well as a fire coral M. dichotoma (Figure 5).
Figure 5.Cladal composition of Symbiodinium population in the collected coral species. Top numerals in parentheses indicate numbers of collected fragments in each species.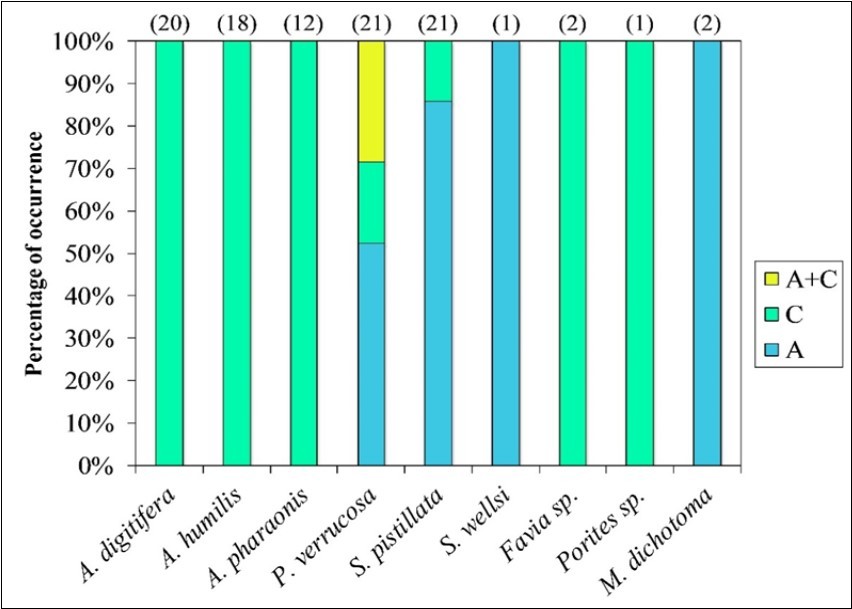
Phylogenetic analysis of Symbiodinium sp. associated with scleractinian corals along wide geographic localities in the Red Sea revealed low diversification in endosymbionts (Figure 6). Nevertheless, eastern coast of the Red Sea showed higher Symbiodinium diversity than western side. Regarding host flexibility, pocilloporid corals hosted more phylogenetically diverse Symbiodinium types. For instance, P. verrucosa harbored Symbiodinium A1, D1a, B1, C15, or C85; while S. pistillata harbored Symbiodinium closely related to A1, F2, C72, or C3. In contrast, Acropora spp. were associated mainly with Symbiodinium C1.
Figure 6.Reconstruction of ITS2 phylogenetic tree between members of Symbiodinium harbored by different taxa of invertebrates in the Red Sea. The phylogenetic distance was inferred by Neighbor-Joining method. Only bootstrap values >50% are shown.
Discussion
In spite of the great concern paid for coral reef ecosystem, Symbiodinium genetic diversity along the Red Sea had been overlooked. So, the current study introduced an overview on Symbiodinium genetic diversity in seven species of scleractinian corals and one species of fire coral along the Egyptian Red Sea coast. The present study revealed that Symbiodinium population associated with Red Sea corals were primarily composed of C and A clades. In general, these two clades were recorded to have a global distribution 20. Among different types of Symbiodinium under each clade, only C1 and A1 types were recorded with the dominance of C1 type comprising 61% of the recorded symbionts in all studied corals. C1 was detected to be the most dominant and widely distributed type because it has the ability to associate with a wide array of host species and a great adaptation with different environmental conditions 21, 22.
Dominance of Symbiodinium C1 was explained by two arguments with the same perception. The first supposed that Symbiodinium C1 type has a generalist nature along wide geographic scales 22. The second proposed that coral-Symbiodinium symbiosis is directed toward specialist-types preference along evolutionary history of coral reefs 21. One of studies suggested that dominance of C1 type in west Pacific had been transferred to western Indian Ocean, and eventually to the Red Sea, before biogeographic partitioning between the two oceans 23. Because northern Red Sea coral reefs are considered recent reefs 24, far from major natural stressors, dominance of C1 type associated with scleractinian corals may be explained on the basis of recent low-specialist symbiosis.
Symbiodinium population at Yanbu was dominated by clade C (69%) followed by clade A (27.3%) while clade D formed only 3.7% of symbionts 11. The current results emphasized the previous mentioned dominance order of clades C and A, and added the endosymbiotic system composed of a combination from the two clades C and A which could be considered as a unique system in Red Sea corals as reported by 20. This is in contrast with corals of the Arabian Gulf where Symbiodinium D represents the dominant clade 11, 25. Predominance of such thermotolerant clade at the Arabian Gulf is associated with thermal tolerance of >33°C 2. However, Symbiodinium D and B that are known to associate with scleractinian corals were not detected.
Compared with high host diversity along the Red Sea, symbiont diversity was low. The same pattern was previously recorded along other reefs at Great Barrier Reef and Indian Ocean 26, 27. The flexibility of P. verrucosa for harboring Symbiodinium A and C was coincident with that recorded in the central Red Sea 28. On the other hand, divergence within phylotype C may explain the dominance of this phylotype in coral reefs of the Red Sea.
Based on coral reproductive strategy, Acropora., Favia and Porites are mainly broadcast spawners and tend to utilize horizontal transmission strategy for Symbiodinium acquisition from surrounding water 10, 29. In contrast, S. pistillata, S. wellsi, and M. dichotoma are principally brooder species and consequently associated with Symbiodinium A by vertical mode of transmission 10. Though, S. pistillata in the Red Sea has the ability to associate with Symbiodinium C 11, 30. Previous studies indicated that colonies of S. pistillata collected from the Red Sea are able to harbor clades A, C, or A+C combination. They stated that 13% of S. pistillata collected from Gulf of Aqaba (17m depth) harbored clade C, while 87% of the colonies (5 and 17m depth) associated with clade A 31, 32. Comparatively, our results revealed that 14% of S. pistillata colonies (mainly collected from surface water at Fanous Reef during winter) were populated with clade C.
It is worth mentioning that the first stages of corals are more flexible than adult stages 33. Simultaneous or individual occurrence of clades A and C in colonies of S. pistillata and P.verrucosa may be interpreted upon assumption that acquisition of Symbiodinium C by colonies that already harbor clade A, may occur by horizontal transmission at later stages during coral ontogeny. A or C clades could be transmitted vertically from parent to juveniles that may maintain cladal structure along life cycle or switch to other clade by horizontal transmission in later stages 34.
At central Red Sea, coral reefs were affected by mass bleaching event at Thuwal during summer 2010 35. They reported that, at some sites, 80-100% of Oculinidae, Agariciidae, and Fungiidae were severely impacted by this event, while bleaching cover among Pocilloporidae, Acroporidae, and Faviidae was 19, 33, and 40%, respectively. More recently, sea surface temperature was raised up to 34°C at the Egyptian coasts of the Red Sea during August 2012. This high temperature anomaly resulted in mass bleaching event in some reef building coral species (Hanafy and Ahmed, unpublished data). Among these, Stylophora, Montipora, Porites, andMillepora were the most affected genera by thermal stress. Symbiodinium A or C harbored by Red Sea corals were considered thermosensitive clades 36. The dominance of these clades observed in the present study may indicate that coral reefs at the Red Sea are endangered by the thermal stress. Accordingly, patterns of temperature anomalies may have dramatic effects on coral reefs diversities and structures at the Red Sea as suggested by 35. However, with little knowledge on Symbiodinium genetic diversity at these levels, we could not generalize our investigation on Red Sea corals. Accordingly, intensive molecular survey with deep resolution on Symbiodinium populations may contribute in understanding of bleaching patterns along the Red Sea. In parallel, future experimental studies are required to derive comprehensive perception about thermal tolerances of these clades and/or types.
Conclusively, the current study emphasizes the need for resolving Symbiodinium genetic diversity along Egyptian coasts of the Red Sea. The level at which this diversity will be resolved is critical for identifying threats to coral reefs at the Red Sea. Identifying Symbiodinium types harbored by different hosts may contribute largely in understanding Symbiodinium dynamics and responses to environmental changes at these reefs.
Acknowledgements
This study was funded by Hurghada Environment Protection and Conservation Association (HEPCA) as a part of MSc scholarship in coral reef restoration.
References
- 1.Stat M, Carter D, Hoegh-Guldberg O. (2006) The evolutionary history ofSymbiodiniumand scleractinian hosts-Symbiosis, diversity, and the effect of climate change. Perspectives in Plant Ecology, Evolution and Systematics 8, 23-43.
- 2.Stat M, R D Gates. (2011) Clade DSymbiodiniumin scleractinian corals: a “nugget” of hope, a selfish opportunist, an ominous sign, or all of the above?. , Journal of Marine Biology,DOI-10.1155/2011/730715 9.
- 3.M A Coffroth, SantosS R. (2005) Genetic diversity of symbiotic dinoflagellates in the genusSymbiodinium. , Protist 156(1), 19-34.
- 4.Sampayo E, Ridgway T, Bongaerts P, Hoegh-Guldberg O. (2008) Bleaching susceptibility and mortality of corals are determined by fine-scale differences in symbiont type. , PNAS 105, 10444-10449.
- 5.K E Fabricius, J C Mieog, P L Colin, Idip D, Oppen M J van. (2004) Identity and diversity of coral endosymbionts (zooxanthellae) from three Palauan reefs with contrasting bleaching, temperature and shading histories. , Molecular Ecology 13, 2445-2458.
- 6.Oppen M J van, A J Mahiny, T J Done. (2005) Geographic distribution of zooxanthella types in three coral species on the Great Barrier Reef sampled after the 2002 bleaching event. , Coral Reefs 24, 482-487.
- 7.A M Jones, Berkelmans R, Oppen M J H van, J C Mieog, Sinclair W. (2008) A community change in the algal endosymbionts of a scleractinian coral following a natural bleaching event: field evidence of acclimatization. , Proceedings of the Royal Society of London Biological Sciences 275, 1359-1365.
- 8.M Y Dosoky, F, M H Hanafy, M I Ahmed. (2014) Distribution ofSymbiodiniumin corals of the Red Sea. , Egypt.International Journal of Environmental Science and Engineering 5, 69-80.
- 9.Barneah O, V M Weis, Perez S, Benayahu Y. (2004) Diversity of dinoflagellate symbionts in Red Sea soft corals: mode of symbiont acquisition matters. Marine Ecology Progress Series 275, 89-95.
- 10.Karako-Lampert S, D J Katcoff, Achituv Y, Dubinsky Z, Stambler N. (2004) Do clades of symbiotic dinoflagellates in scleractinian corals of the Gulf of Eilat (Red Sea) differ from those of other coral reefs?. , Journal of Experimental Marine Biology and Ecology 311, 301-314.
- 11.A C Baker, Jones S H I, T S Lee. (2005) Symbiont diversity in Arabian corals and its relation to patterns of contemporary and historical environmental stress. In. Proceedings of an international symposium on the 24-36.
- 12.PERSGAGEF. (1990) Coral reefs in the Red Sea and Gulf of Aden. Surveys. PERSGA Technical Series No. 7. PERSGA, Jeddah to 137.
- 13.Veron J E N. (2000) Corals of the World. Australia: Australian Institute of Marine Science and CRRQldPtyLtd.
- 14.M R Gaither, Szabó Z, M W Crepeau, C E Bird, R J Toonen. (2011) Preservation of corals in salt-saturated DMSO buffer is superior to ethanol for PCR experiments. , Coral Reefs 30, 329-333.
- 15.M A Coffroth, H R Lasker, M E Diamond, J A Bruenn, Bermingham E. (1992) DNA fingerprints of a gorgonian coral: a method for detecting clonal structure in a vegetative species. , Marine Biology 114, 317-325.
- 16.Rowan R, D A Powers. (1991) Molecular genetic identification of symbiotic dinoflagellates (zooxanthellae). Marine Ecology Progress Series 71, 65-73.
- 17.T C LaJeunesse, R K Trench. (2000) Biogeography of two species ofSymbiodinium(Freudenthal) inhabiting the intertidal sea anemoneAnthopleuraelegantissima(Brandt). , Biological Bulletin 199, 126-134.
- 18.N 1 Saitou, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. , Mol. Biol. Evol 4(4), 406-425.
- 19.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution. 30, 2725-2729.
- 20.M J vanOppen, A C Baker, M A Coffroth, B L Willis. (2009) Bleaching resistance and the role of algal endosymbionts. In: M.J.H. Van Oppen and J.M.Lough, Eds.,Coral bleaching:patterns, processes, causes, and consequences. , Berlin:Springer 83-102.
- 21.T C LaJeunesse, D J Thornhill, E F Cox, F G Stanton, W K Fitt. (2004) High diversity and host specificity observed among symbiotic dinoflagellates in reef coral communities from Hawaii. Coral reefs. 23, 596-603.
- 22.Y L Chan, Pochon X, M A Fisher, Wagner D, G T Concepcion. (2009) Generalist dinoflagellate endosymbionts and host genotype diversity detected from mesophotic (67-100 m depths) coralLeptoseris. , BMC Ecology 9, 21.
- 23.T C LaJeunesse. (2005) Species" radiations of symbiotic dinoflagellates in the Atlantic and Indo-Pacific since the Miocene-Pliocene transition. , Molecular Biology and Evolution 22, 570-581.
- 24.Shaked Y, Genin A. (2011) Red Sea and Gulf Of Aqaba. In:D. Hopley, Ed.,Encyclopedia of modern coral reefs.Springer. 839-843.
- 25.P G Mostafavi, Fatemi S M R, M H Shahhosseiny, Hoegh-Guldberg O, Loh W K W. (2007) Predominance of clade DSymbiodiniumin shallow-water reef-building corals off Kish and Larak Islands (Persian Gulf. , Iran), Marine Biology 153, 25-34.
- 26.Hoegh-Guldberg R, Schmidt O, W G. (2003) Low symbiont diversity in southern Great Barrier Reef corals, relative to those of the Caribbean. Limnology and. Oceanography,48 2046-2054.
- 27.S Y Yang, Keshavmurthy S, Obura D, Sheppard C R C, Visram S. (2012) Diversity and distribution ofSymbiodiniumassociated with seven common coral species in the Chagos Archipelago, central Indian Ocean. PLoS One,7(5), e35836
- 28.Sawall Y, Al-Sofyani A, Banguera-Hinestroza E, C R Voolstra. (2014) Spatio-temporal analyses ofSymbiodiniumphysiology of the coralPocilloporaverrucosaalong large-scale nutrient and temperature ggradients in the Red Sea. , PLoS One 9(8), 103179.
- 29.M H Hanafy, M A Aamer, Habib M, A B Rouphael, A H Baird. (2010) Synchronous reproduction of corals in the Red Sea. , Coral reefs 29, 119-124.
- 30.Barneah O, Brickner I, Hooge M, V M Weis, T C LaJeunesse. (2007) Three party symbiosis: acoelomorph worms, corals and unicellular algal symbionts in Eilat (Red Sea). , Marine Biology 151, 1215-1223.
- 31.Keshavmurthy S, S Y Yang, Alamaru A, Y, Pichon M. (2013) DNA barcoding reveals the coral “laboratory-rat”,Stylophorapistillataencompasses multiple identities. Scientific Reports,DOI-10.1038/srep01520. 3, 1520.
- 32.Lampert‐Karako S, Stambler N, D J Katcoff, Achituv Y, Dubinsky Z. (2008) Effects of depth and eutrophication on the zooxanthella clades ofStylophorapistillatafrom the Gulf of Eilat (Red Sea). , Aquatic Conservation of Marine and Freshwater Ecosystems 18, 1039-1045.
- 33.S V Vollmer, A C Baker, M A Coffroth, C D Harvell, Medina M. (2013) Understanding the coral holobiont through science. 173-186.
- 34.K A Byler, Carmi-Veal M, Fine M, T L Goulet. (2013) Multiple symbiont acquisition strategies as an adaptive mechanism in the coralStylophorapistillata. , PLoS One 8(3), 59596.
