The Use of Metabolomic Tool in Assessing Environmental Exposure
Abstract
The impact of the environment on the development of non-communicable chronic diseases has gained prominence in recent years. In this context, a new chemical exposure assessment strategy is needed that is capable of revealing multiple exposures, as well as reflecting the cumulative interaction between such environmental contaminants in the biological system. From this perspective, metabolomics emerges as a promising tool in this field of knowledge, since it is able to identify changes in metabolism and/or gene expression resulting from exposure to environmental factors. The aim of this study was to describe important concepts, as well as the steps that permeate the metabolomics analysis, and also to present some relevant works with the application of metabolomics in the assessment of chemical exposure. A literature review showed a significant increase in the use of metabolomics in environmental toxicology in recent years. This increase is mainly due to advances in analytical techniques and the improvement of data processing tools. However, this field of investigation remains little explored, especially with regard to the study of toxicity associated with chronic exposure to low levels of chemical agents. Thus, it is urgent that omic biomarkers can be used as a tool for decision-making, especially with a view to protecting, diagnosing and recovering human health.
Author Contributions
Academic Editor: Sasho Stoleski, Institute of Occupational Health of R. Macedonia, WHO CC and Ga2len CC.
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2021 Michele Polyana Rocha Mendes, et al.
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
The population is constantly exposed to several chemicals from sources such as food and water, air breathing, medicines and personal care products; some of which do not have their toxicity fully known. Several recent studies have highlighted the importance of associating environmental exposure to chemicals and development of chronic diseases acknowledged as the main global causes of death, such as cardiovascular and degenerative diseases, diabetes and cancer1.
Chronic diseases can result from gene-environment interaction, since they are caused by both epigenetic and gene expression changes, which are conditioned by lifestyle and acquired infections. This concept highlights the role of the environment, which is understood as a non-genetic cause of chronic diseases 2. The role played by environmental factors in disease pathogenesis has been the focal point of several studies seeking to demonstrate the real impact of the environment on human health 3, 4, 5, 6, 7, 8, 9, 10.
Interaction of environmental factors — such as chemical exposure, radiation, lifestyle, exercising, occupation, diet and obesity — with the human organism changes qualitatively and quantitatively over time 4, 5, 11. As the consequence of such a variation, assessing environmental impacts on health became even more challenging than it used to be.
Wild (2005)3 proposed the concept of exposome — total of environmental exposures from conception to death — for full chronic disease pathogenesis assessment. Genetic research, such as the human genome mapping from the 1980s-90s, contributed to develop genotyping methods likes PCR and microarray analysis to assess the disease-gene relationship. However, exposome analysis has become a reality through omics technologies such as metabolomics, which is the study of metabolites that carry information about chemically induced molecular mechanisms in cells and tissues. This study can be quickly and accurately performed through high-performance analytical methods like chromatography and mass spectrometry 12, 13, 14, 15, 16, 17.
Although most toxic effects of chemicals are well known, little is known about metabolic responses to environmental exposure in the general population. Metabolomics has been an important tool to improve comprehensive analysis about microbiota-xenobiotic interactions; it has also been an alternative to epidemiological studies on the toxicity of environment chemicals. Exposure to such chemicals is usually mild — due to low chemical concentrations in the environment —, yet prolonged 6, 18, 19, 20, 21, 22, 23, 24. The advantages of assessing the metabolome include analysis of genetic factors, endogenous metabolites and environmental exposures 25. Accordingly, advances in analytical methods, data processing, quality control and multivariate statistical and chemometric analyses have improved metabolomics’ accessibility to environmental-epidemiology studies 26, 27.
The present article introduces the metabolomics approach and its potential as tool to help understanding molecular mechanisms induced by environmental toxin exposure. Understanding this issue can help finding potential early and sensitive biomarkers to assess overall health risks in the population.
Metabolomics - Perspectives For Assessing Environmental Exposure To Chemical Agents
The term metabolome, similarly to genome and proteome, means the set of all metabolites (lipids, carbohydrates, vitamins, fatty acids, secondary metabolites, signaling molecules, hormones and others) presenting molecular mass up to 1500 daltons in a given biological sample 17, 25, 27. Thus, it is worth noticing that metabolome structure is divided into segments, i.e., the profile of plasma metabolite differs from that of urine, which differs from that of saliva, and so on. Nicholson et al., (2012)26 claim that humans have more than 500 different metabolites, since approximately 500 different cell types are known to produce several metabolites. The Human Metabolome Database - HMDB (2018) recorded approximately 114.500 identified metabolites28. This amount is relatively smaller than that of genes and proteins, which makes metabolomics even more attractiv e as scientific field 29.
Although metabolomics and metabonomics are commonly regarded as synonyms, there is a subtle difference between them: metabonomics is the study of metabolite interactions in a complex system over time; it was introduced by Nicholson; Lindon and Holmes (1999)30 as the quantitative measurement taken from the dynamic multiparametric metabolic response of living systems to pathophysiological stimuli or genetic modification. On the other hand, metabolomics was defined by Fiehn (2001)31 as the comprehensive analysis of all metabolites in a given biological system under certain conditions. The first manuscript using the word metabolome was published in 1998; since then, an increasing number of publications on this topic have been reported. The Figure 1 shows the comprehensiveness of metabolomics studies and chemical exposure described by publications from the last 11 years.
Figure 1.Number of publications located in Pubmed on metabolomics and metabolomics approach using analytical platforms - LC-MS, GC-MS, NMR and CE-MS in toxicological studies in the period 2009-2020.
Exposure to environmental toxins leads to changes in metabolism and/or gene expression (deletion or overexpression). Such an alteration may indicate a specific pattern called “metabolic signature”, which has the ability to signal this exposure 16. Several studies on metabolomics and environmental exposure to toxins have been proposed based on this concept.
Metabolomics has been proven to be an efficient strategy to assess exposure to chemical agents. It showed great potential for studying the toxicity of chemical substances 32, since the metabolite profile can provide a broad view of the physiology of a given organism 31. Accordingly, Holmes et al., (2007)33 proposed the term “xenometabolomics” to characterize the profile of metabolites derived from the biotransformation of xenobiotics.
Two metabolomics-study approaches have been used to detect metabolites: untargeted and targeted metabolomics25. Untarget metabolomics is the comprehensive analysis of all measurable metabolites, and it has been conducted to understand environmental toxin mechanisms 18, 34, 35, 36, 37, 38, 39. Biomarker candidates detected in the untargeted analysis can be validated by targeted metabolomics. Targeted metabolome analysis is based on biological hypothesis and quantitatively investigates differential gene expression from pre-selected metabolite candidates. Metabolites presenting differential gene expression associated with toxin exposure and/or disease must be subjected to pre-clinical and clinical validation, as elucidated by Figure 240.
Figure 2.Stages of the discovery of new biomarkers in metabolomic
Pre-clinical validation can be done through in vitro testing by comparing control and sickle cell lines of pre-selected metabolite(s) or by adding metabolites to a cell line in order to check whether they can induce the development of pathological phenotypes. Next, clinical validation through biomarker detection in real biological samples must be performed in order to determine biomarker robustness and reproducibility 41. Analysis of such data reveals that the untargeted approach and GC/LC-MS-based metabolomics are the most common me tabolomics methods. However, there is no prevalent study design — animal, cell culture and real biological samples have all been applied to metabolomics studies.
Yet, the robustness and reproducibility of metabolite profile analysis are impaired by the following factors: highly variable metabolite quality and quantity; wide range of metabolite physicochemical properties 42. Capillary electrophoresis, chromatography methods and nuclear magnetic resonance (NMR) — all associated with mass spectrometry — have been widely applied to metabolomics studies. However, none of these analytical methods is able to fully detect and quantify metabolites 25, 42, 43. Alternatively, several procedures and mixed methods, such as GC/LC-MS-based metabolomics data, can be performed to broaden both mass spectrometry and physicochemical property coverage 44, 45.
Biomarkers traditionally applied to environmental exposure assessments are mostly related to recent exposure rather than to adverse health effects, which are most likely related to health risks; therefore, environmental risks are underestimated 16. Broad assessment strategies able to detect several toxin exposures and to reveal the cumulative interaction (synergism and antagonism) among them are key in assessing the impact of environmental toxin exposure on human health.
Metabolomics is aimed at overcoming such limitations and fostering comprehensive analysis strategies, since it enables a new perspective on the first molecular mechanisms triggered by environmental toxin exposure 46. Changes in the metabolite profile caused by exposure to toxins interacting with the biological system assumingly indicate toxicity biomarkers that can be applied to future studies 47.
Therefore, studies point towards advances in environmental toxicology and to a new outlook for assessing environmental contamination risks: “Omic” biomarker assessment should be advanced enough to detect both environmental toxins and their adverse health effects (disease biomarkers) in a single diagnosis. Biomarker data analysis should become a tool for clinical decision-making in order to protect, diagnose and recover human health.
Metabonomics: Analytical Aspects
Metabolomics studies require rigorous experimental planning due to the wide range of factors leading to biased and/or inaccurate results. Dudzik et al., (2018)48 state that a good experimental design must cover different variables to ensure both impartial sample analysis and solutions to specific biological issues. The literature presents several pre-analytical biological factors, both intrinsic and extrinsic, affecting the basal metabolic rate. Intrinsic factors include age, sex, genotype, ethnicity, chronobiology, body mass index (BMI) and sample volume, whereas extrinsic factors include eating habits, stress, physical activity, microbiota, environmental toxin exposure and consumption of drugs, alcohol and tobacco 49, 50, 51, 52, 53, 54, 55, 56, 57. Individuals can be evenly distributed according to weight, age and sex across all different groups in order to limit the effect of the aforementioned factors on statistical analysis57, 58. Moreover, results should be normalized to minimize inaccuracy caused by both metabolite concentration variability and method detection limit. Normalization is particularly important for urine samples, since urine metabolome is influenced by urinary volume and renal clearance. The following urinary biomarkers should be normalized for best results: abundant metabolic signals in each sample, urinary volume, creatinine levels and urine osmolality. Creatinine normalization is a common method, since the impact of external factors on urine concentration can be determined by measuring physiological substances in urine samples, such as creatinine. In other words, urinary creatinine levels determine urinary concentration. Accordingly, the concentration of a given metabolite must be divided by its creatinine levels for normalization purposes 59, 60.
Studies must be well controlled through experimental design in order to minimize analytical errors and avoid result bias 42, 58. Therefore, biological issue, study population and biological matrices of choice must be well known to ensure the following: sample homogeneity and representativeness; proper performance of analytical methods, data processing and statistical analysis 48.
Metabolome analysis uses one or more combined analytical methods and bioinformatics tools to assess research results. This analysis is key in understanding the biochemical mechanisms exposed to environmental toxins, since it provides qualitative and semi-quantitative data from affected metabolites through untargeted and targeted approaches 61. These approaches have similar workflows, since experimental procedure standardization and result publication are high priority for result comparisons and further studies. Accordingly, the Metabolomics Standards Initiative (MSI) (http://www.metabolomics-msi.org/), conceived in 2005, defined the procedures to be adopted in studies and publications to achieve such a harmonization. Both targeted and untargeted technologies involve the following steps: defining the biological issue of interest, experimental design, biological sample collection and preparation; data acquisition and analysis; biological interpretation of results 17, 62, 63.
Both untargeted and targeted metabolomics workflows differ in the first step and in the required sample preparation: the untargeted workflow starts by defining the biological issue of interest, which represents the question(s) that must be answered at the end of the study, whereas targeted metabolomics starts with a biochemical hypothesis and focuses the quantitative metabolite analysis based on one or more previously defined metabolic pathways of interest 17, 63, as shown in Figure 3
Figure 3.Workflow summarized in untargeted and target metabolomics.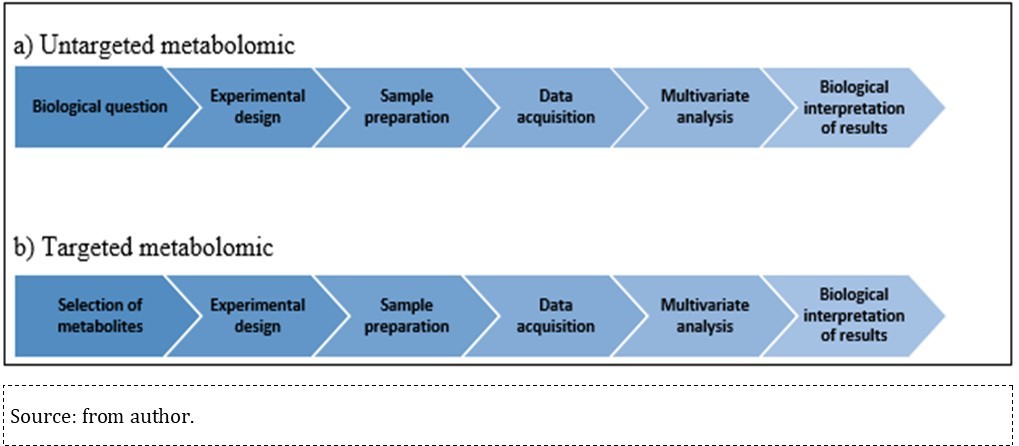
Experimental Design
Experimental design requires systematic planning and the performance of procedures such as defining the biological matrix of interest, sample number per group, sample collection and storage. Plasma and urine are the most common samples, whereas other samples such as saliva, sweat, exhaled air, cerebrospinal fluid and seminal fluid are the least common ones 64. Samples should be collected with due attention, since several lifestyle-related pre-analytical factors can affect the metabolite profile. Thus, data such as collection time, need of fasting, and anticoagulant and preservative use should be previously defined. Slupsky et al., (2007)57 demonstrated that eating habits can affect urinary metabolite concentration. Therefore, they recommend that urine should be collected in early morning hours to avoid variations. Urinary metabolite profile findings can be altered by factors such as collection and fasting periods, centrifugation conditions, filtration, food additives (e.g. sodium azide), normalizing procedures and repeated freeze-thaw cycles. Plasma is assumingly more stable than serum, whereas serum can provide greater sensitivity than plasma 65. Thus, samples must be chosen based on the required sensitivity-reliability analysis results.
Yet, control groups should be selected to allow comparisons between their results and those of exposed groups. Although this approach has been traditionally applied to toxicological studies, it has been adapted: the exposed group is classified based on exposure intensity and comparison is made between the lowest and highest exposure groups 19, 37. Thus, some factors must be considered prior to analysis procedures, such as whether the result comparison of studies with different approaches is valid and whether it should be performed with caution. Therefore, assessments involving discussion and comparison of both traditional and modern result comparison approaches should be conducted in order to guide the scientific community and ensure result reproducibility.
Sample Collection, Storage and Preparation
Metabolic processes must be stopped after sample collection in order to preserve samples — this procedure is known as metabolic quenching. This step is normally accomplished by adding organic solvent to the samples and immediately cooling or freezing them with dry ice or liquid nitrogen 17, 66. Samples must be stored at low temperatures, preferably at -80ºC, in order to minimize potential bacterial contamination and sample degradation when instrumental analysis is performed long after sample collection 48. Results of studies assessing metabolomic profile stability in different biological samples have demonstrated that metabolite stability depends on several factors, as follows: time period between collection, processing and storage; number of freeze-thaw cycles; storage time and temperature. Pinto et al., (2014)67 reported a 2.5-year stability of the plasma metabolomic profile, whereas the literature review conducted by Stevens et al., (2019)65 reported a 26-month stability for urinary metabolite samples stored at room temperature (-25 ºC). Semren et al. (2018)68 reported insignificant decrease in urinary metabolites stored at -80 ºC for 6 months.
Metabolite sample preparation is one of the most important steps of the procedure. It should be based on desired approaches, biological matrices and analytical methods of interest. Sample preparation for untargeted metabolomics must be minimal in order to avoid metabolite loss during the process, since the comprehensive metabolic analysis is the aim of the preparation. This step usually involves the following procedures: deproteinization to minimize matrix effects and clogging of ionic compounds 63; solvent delipidation; refrigerated centrifugation to separate unnecessary particles such as cells, waste and high-molecular-weight metabolites; solvent dilution and precipitation 56, 68, 69. Next, liquid-liquid (LLE) and solid phase (SPE) extractions are performed. Urine samples require previous urease preparation to minimize pre-analytical and analytical factors of samples analyzed through the GC-MS method. Urea can interfere with derivatization and lead to incomplete reactions, since it is the most abundant physiological metabolite in urine. Accordingly, concentrated urea can overload chromatographic columns and cause chromatographic peak distortions due to co-elution. Therefore, it is recommended to add 30 µL of urease to each 100 µL of urine and incubate at 37 ºC for 30 to 60 minutes 24, 70, 71, 72. On the other hand, targeted metabolomics is based on selective sample preparation, since it aims at quali-quantitative analysis of one or more metabolites with similar physicochemical properties. Therefore, targeted metabolomics requires both highly selective extraction methods and preconcentration and clean-up procedures to eliminate distortions 64.
Derivatization must be performed in order to improve metabolome coverage through the GC-MS method, since most natural metabolites contain non-volatile polar functional groups (organic acids, amino acids, monosaccharides, disaccharides and steroids). Thus, methoxymination reactions followed by silylation — the most common reactions — allow improving analyte properties by making analytes volatile and thermally stable70, 73, 74. Instrumental analysis through capillary electrophoresis and nuclear magnetic resonance methods requires full organic solvent evaporation, subsequent waste resuspension in water and dilution of extracts in deuterated solvents 17.
Instrumental Analysis – Data Collection
A fraction of the sample is collected at the end of the preparation step and subjected to analytical platforms for the next step: data acquisition. It is worth noticing that the aim of metabolome analysis is the study of all metabolites (untargeted) of molecular weight up to 1500 Da or the study of part of them (targeted) - these analytes have different concentrations and great chemical diversity. Therefore, metabolites should be assessed through multiplatform analyses, as they can cover the entire metabolome spectrum. Combined chromatography and mass spectrometry (LC-MS and GC -MS) have been the most widely performed metabolomic methods in the last decade. Nuclear magnetic resonance (NMR) and capillary electrophoresis combined with mass spectrometry (CE-MS) are also reported in the literature [75,76,77].
Combined chromatography and mass spectrometry considerably improves qualitative-quantitative analysis of complex biological samples, since the analytes of a given sample can be separated in the chromatograph and moved into the mass spectrometer where they will be ionized, identified and quantified. When these analytical methods are combined, molecular entities can firstly be identified through chromatographic retention time and accurate mass measurement77. Moreover, fragmentation pattern can be the parameter to identify molecular entities when data are obtained through MS-MS (MS tandem)20.
Mass spectrometry plays a leading role in metabolomics research due to its high analytical sensitivity and specificity, which generate ions to be separated in the mass spectrometer based on their mass (m) to charge (z) ratio (m/z). Next, the generated mass spectrum is compared to spectral databases or patterns of known substances77,78. Mass analyzers should be able to analyze a wide range of masses at high resolution, i.e., they should distinguish two m/z peaks differing by only a single atomic mass unit for metabolome analysis78. Hybrid mass spectrometers, which use more than one mass analyzer, such as triple quadrupole, and quadrupole time-of-flight (Q-TOF-MS) analyzers, improve the resolution and precision of ion mass measurements. Therefore, they are the most appropriate analyzers for metabolome analysis44, 79, 80. Different ionization processes must be performed for the coupling of mass spectrometers, due to structural differences between LC and GC methods. GC-MS uses Electron Ionization (EI) and chemical ionization (CI), whereas LC uses Electrospray Ionization (ESI) — the most common LC method —, Atmospheric Pressure Ionization (API) and Atmospheric Pressure Chemical Ionization (APCI)81.
Selection of analytical ionization methods must be based on desired polarity and molecular mass range. GC-MS is assumingly the most suitable method for low-polar, low-molecular-weight compounds, whereas LC-MS-ESI is the most suitable for highly polar, high-molecular-weight compounds. Furthermore, these analytical methods are highly complementary, which explains why they are combined in order to cover the entire metabolome 82.
Yet, both methods have pros and cons. GC-MS is a very robust method, whose sensitivity, selectivity and reproducibility are suitable for simultaneous metabolite analysis 61, 83, 84. Moreover, it stands out for its retention-time reproducibility and its low-cost, comprehensive databases. However, it has a limitation: lack of comprehensive MS-MS fragmentation data 45. Comprehensive GC-MS-based metabolomics protocols on sample preparation and analytical conditions have been described by Fiehn (2017)85, Garcia and Barbas (2011)70 and Mastrangelo et al., (2015)83 .
The LC-MS method is particularly important to solve complex matrices 77. It is also suitable for metabolomics studies due to its great versatility (variety of mobile and stationary phases) 17, 44, 77, 86. Hydrophilic interaction liquid chromatography (HILIC) has been reported for polar metabolite analysis, polyhydroxy metabolite analysis and reverse phase chromatography (relatively non-polar metabolite analysis) 38, 77, 86. The advantages of this method are high precision and resolution; analytical sensitivity and specificity. Furthermore, LC-MS requires simplified sample preparation and is applicable to complex polar and nonpolar mixtures. The only disadvantage of LC-MS lies on the high cost of replacing consumables by highly pure and compatible mobile phases 45.
Procedures to assess data acquisition quality should be performed in sequence analysis. Such a quality is assessed through quality management processes like Quality Assurance (QA) and Quality Control (QC) 48,87,88. QA involves all the planned and systematic activities implemented in the pre-analytical phase to ensure that preset quality requirements will be fulfilled by subsequent analytical processes. Currently, there are no recommended guidelines for the QA process. QC can be defined as the operational techniques and activities used to measure and report quality requirements during and after data acquisition. Quality control samples can be collected by solutions presenting authentic chemical standards, samples pooled with equal aliquots of biospecimens from different biological sources or samples pooled with equal volumes 48,87,88. Pooling samples are commonly used in QC because they show the complete metabolite structure of all biological samples in a given study. Thus, significant quality control deviations indicate analytical variability resulting from sample preparation or instrumental analysis phases. This strategy has been applied to assess the stability and performance of control systems 43, 89. Dudzik et al., (2018)48 recommend intermittent QC analysis after every 5 to 10 samples throughout the analytical run and state that variation coefficients lower than 30% in each peak should be considered acceptable. Graphical representation of QC results through PCA models is a practical way to assess control stability, since intense clustering indicates good accuracy 48, 90.
However, the 2017 questionnaire developed by the Metabolomics Society Data Quality Task Group (DQTG), assigned to 97 employees from 84 metabolomics companies, evidenced that there is no consensus on QC procedures and decision-making regarding data quality in the international community87. Thus, further research should be encouraged to foster the development of reliable QA and QC strategies. These strategies should be commonly applied to metabolomics workflows in order to improve the overall quality of results.
Data Analysis and Interpretation
Metabolome analysis captures a huge amount of exceedingly complex biochemical data (disease or chemical exposure) that makes manual inspection impractical. Thus, bioinformatics tools are indispensable for data processing. Untargeted metabolomics data analysis is laborious and involves several steps, such as: converting raw data into datasets; data collection and alignment; retention time and baseline correction, spectral deconvolution, data normalization; metabolite identification17. There are several software for analyzing data collected through combined analytical methods, such as XCMS®, AMDIS®, Mass Professional Profile® and MetAlign® 25, 70, 85, 86, 91, 92. On the other hand, targeted metabolomics data analysis is focused on the quantitative assessment of previously selected metabolites 17.
The normalized dataset is subjected to multivariate chemometric analysis through unsupervised methods, such as principal component analysis (PCA), and supervised methods, such as partial least squares-discriminant analysis (PLS-DA) and orthogonal projections to latent structures discriminant analysis (OPLS-DA). These methods have clear purposes: PCA preliminarily identifies sample clusters to reduce the dimensionality of large datasets, in addition to allowing the identification of outliers. On the other hand, PLS-DA is a regression method that reveals the variables mostly causing result variability and identifies differences between assessed groups. Finally, OPLS-DA identifies potential biomarkers 17,93. Several tools are available for multivariate data analysis, including: MetaboAnalyst® 94, 95 - a set of free online tools that can be used in both pre-processing and multi- and univariate statistical analysis and metadata interpretation; SIMCA-P® paid software developed by Umetrics; Matlab® 17.
Univariate statistical tests such as Student’s t-test, Analysis of Variance (ANOVA) and Mann-Whitney U test are commonly performed to assess the statistical significance of each peak (molecular entity) 17, 36, 74, 92.
Commercial databases such as NIST, HMDB, Blood Exposome and METLIN have been accessed to help identifying metabolites 86, 91. However, it should be noticed that most metabolites are not detectable. Hounoum et al. (2016)86state that approximately 25% of them are identified by attempt, since many metabolites are not added to metabolite databases and repositories. To harmonize metabolomics studies, in 2005, the Society of Metabolomics created the Metabolomics Standards Initiative (MSI), which advocates the standardization of the entire experimental procedure, including 4 confidence levels for identification of metabolites 122.
Yet, biological data interpretation is as important as the previous steps. This procedure is performed by placing the identified metabolites into a metabolic pathway or network in order to identify their variation rate and compare it to that of the control group. This step aims at finding the answer to the biological question established at the beginning of the workflow to ensure meaningful data interpretation17,92. Pathway mapping, visualization and enrichment analyses are common and accessible tools in databases86 such as Kyoto Encyclopedia of Genes and Genomes (KEGG)96, MetaCyc 97, MarVis-Pathway98 and RaMP99used to find the mechanistic view of identified metabolites100. Quantification of identified metabolites is a common procedure of targeted metabolomics 17.
Piovezan (2014)90 states that metabolomics results should be systematically reviewed at the end of each workflow step. Analytical validation is performed through the following procedures: visual inspection of all analyses and of LC analysis’ pressure curves; evaluation of reprocessed data according to the number of molecules extracted per sample, their total signals and QC results. On the other hand, chemometric models are evaluated through statistical tests such as R2 and Q2, or through cross-validation and permutation testing. Finally, biological validation must be performed in targeted metabolomics studies90.
Metabolomics Application to Environmental Toxicology
When mixed chemicals (xenobiotics) enter the body, they can either act in single metabolic pathways or overlap in common metabolic pathways, according to Vineis (2018)101. Thus, estimating of low-level chronic chemical exposure risks through conventional epidemiological studies is insufficient, since it cannot cover all the complexity of cumulative exposure effects. In view of this limitation, new strategies for cumulative risk assessment should be developed in order to provide real estimate of such risks102. Therefore, new technologies have been developed to provide new methods to assess cumulative exposure and its health risks. Metabolomics has been used to assess cellular disorders induced by chemical exposure, estimate disease risk and detect new biomarkers24, 40, since environmental pollutants lead to early biochemical changes in metabolite levels, which can reveal disease onset and prognosis. These changes in metabolite levels are called metabolomic signatures or metabolomic profiles. Thus, metabolomics allows qualitative and quantitative analysis of the metabolomic profile in body fluids, before the onset of clinical symptoms. Moreover, molecular profiling can be used to identify exposure, diagnosis and/or prognosis biomarkers103. Such a comprehensiveness significantly increases the opportunities for toxicological and environmental health actions. It also encourages discussions on a new biomarker classification, whose metabolomic profile (metabolomics biomarker) embodies the role of exposure and/or effect biomarker.
In view of the high risk of developing chronic diseases due to environmental chemical exposure16, 102, the lack of publications on metabolomics approach to environmental chemical exposure reveals that this knowledge field has been little explored. Therefore, further research should be encouraged in order to unveil the poisoning pathways of chemicals and develop measures to cure, protect and recover the health of exposed individuals. Some relevant studies proving metabolomics’ potential to assess environmental chemical exposure are described below.
Studies carried out in China have assessed exposure to arsenic19,37— an environmental threat to human health, since exposure to it can lead to several diseases, such as cancer, cardiovascular diseases and peripheral neuropathy104. Zhang et al., (2014)19 assessed the urinary metabolome from Chinese men environmentally exposed to arsenic through the HPLC/ Q-TOF-MS method. Individuals were classified according to urinary arsenic level and metabolite profile (comparison between highest and lowest exposure groups). Untargeted metabolomics detected five dose-dependent arsenic, differentially expressed metabolites (testosterone, guanine, hippuric acid, N1-acetyl-N2-formyl-5-methoxykynuramine (AFMK) and serine). Based on such results, the authors stated that endocrine disruption and oxidative stress are the likely causes of arsenic poisoning. They assessed the biomarker potential (selectivity and specificity) of candidates through the ROC (Receiver Operating Characteristic) curve. Results showed that testosterone, guanine, hippuric acid and the combination of them all were gene signatures of arsenic-induced metabolic disturbances. AUC (Area Under the Curve) measures the overall potential of a test to discriminate whether a specific condition is present or not. AUC biomarker values greater than 0.85 are usually considered acceptable for clinical application 19, 105.
Chemical exposure during pregnancy has attracted the attention of the scientific community. The effects of arsenic on pregnant women’s health have also been assessed. The urinary metabolite profile of Chinese pregnant women was analyzed for the first time through the UPLC/Q-TOF-MS analytical method in order to identify the adverse effects of low-level arsenic exposure. The urinary arsenic of all participants was measured and used as reference to group urine samples according to exposure level. Metabolome analysis aimed at assessing metabolite profile and comparing the lowest and highest exposure groups. Results indicated the detection of 9 significantly altered metabolites. The predictive capacity of these metabolites was assessed through the ROC curve, which demonstrated that all 9 metabolites had enough predictive capacity (AUC > 0.8) to be considered potential biomarkers 37. Metabolic disturbances in pregnant women and their fetuses resulting from exposure to perfluoroalkyl substances (PFASs), persistent organic pollutants, were reported by Li et al (2021) 106. The authors identified that exposure to PFAS led to changes in steroid hormone biosynthesis and acid metabolism fatty acids, vitamins, amino acids and lipids. The pathway enrichment analysis showed that 3-fatty acid metabolism and retinol were significantly correlated with exposure to PFAS in maternal blood, and sterol metabolism was found to be significant in both maternal serum and umbilical cord serum. In addition, serum metabolomics from 102 Chinese pregnant women revealed disruption of thyroid hormone metabolism and glyceraldehyde metabolism. These disturbances have been associated with birth impacts resulting from prenatal exposure to pesticides such as mecarbam and β-hexachlorocyclohexane (β-HCH) 107.
A recent study conducted by Wang et al. (2018) 104 assessed metabolite changes induced by combined exposure to polycyclic aromatic hydrocarbons (PAHs) and short-chain chlorinated paraffins (SCCPs) in Hep-G2 cells. Significant changes have been detected in several metabolic pathways, such as phospholipid metabolism, fatty acids, tricarboxylic acid cycle, glycolysis and purine metabolism. Wang et al. (2018)104 highlighted that lipid metabolism disorder was induced by combined exposure to PAHs and SCCPs. Accordingly, this finding requires further research due to its severe impact on human health.
PAHs are organic contaminants having at least two condensed or fused aromatic ring structures. They are ubiquitous in the environment and stem from the incomplete combustion of organic compounds. PAHs are present as vapors or adhered to particulate matter. Exposure to PAHs can cause cancer, teratogenesis and mutations20, 105, 106. The harmful effects of chronic exposure to PAHs were assessed by Wang et al., (2015)107 in 566 Chinese volunteers — children and elderly individuals. Untargeted urinary metabolome analysis detected 18 discriminating metabolites between the exposed group and the control group, of which dodecadienylcarnitine and 1-hydroxyphenanthrene (1-OHPh) were potential biomarkers to assess the exposure of the general population to PAHs. The authors suggest that these metabolic changes are caused by PAH-induced oxidative stress, and that supplemental antioxidants should be administered to neutralize or reverse the harmful effects of these hydrocarbons.
Heavy metals are chemicals of great public health concern. Cadmium metal comes from both natural sources and human environmental impacts, and it is suitable for a wide variety of industrial applications. This metal is a human carcinogen and exposure to high doses of it can lead to adverse health effects such as itai-itai disease, which causes osteomalacia, osteoporosis and renal tubular dysfunction 108, 109. Cases of death from cadmium poisoning have been reported in Thailand and Japan 24, 109, 110. Contaminated individuals living in Mae Sot City, Tak province, Thailand were selected for research on the metabolite profile of individuals intoxicated by cadmium. The study aimed at identifying cadmium biomarkers (urinary cadmium) and the urinary metabolite profile of intoxicated and healthy patients (control) through GC-MS analysis. Results indicated that urinary citrate and myo-inositol may be potential biomarkers for Thais exposed to Cd. Moreover, urinary citrate has also been proven to aid in early nephrolithiasis diagnosis and prevention 24. Cadmium-induced nephrotoxicity was investigated in another recent study, conducted by Zeng et al (2021) 116. The urinary metabolic profile of 149 individuals (99 women and 45 men) residing in areas contaminated with different levels of cadmium. The metabolic profile showed that exposure to this metal caused alternations in the creatine pathway, amino acid metabolism, especially tryptophan metabolism, aminoacyl-tRNA biosynthesis and purine metabolism, regardless of gender. Therefore, these findings should be explored to identify early-effect biomarkers that can be used to predict risk and prevent cadmium-induced nephrotoxicity.
Another environmental contaminant of toxicological importance is particulate matter (PM), which is among the main air pollutants. PM consists of liquid or solid particles suspended in the air, whose composition is quite heterogeneous and includes toxic substances such as polycyclic aromatic hydrocarbons (PAHs) and metals (Pb, Hg, V, Cd, Cr and others) 117, 118. Several studies have shown the association between prolonged exposure to PM, especially PM2.5 (<2.5 μm in aerodynamic diameter) and an increased risk of cardiovascular and metabolic diseases 119,120. Chu et al. (2021)121 performed a prospective cohort study to investigate metabolic changes in plasma of 78 Chinese university students, 36 men and 42 women; exposed to PM2.5 and PM10 (mean concentration of 53 μg/m3 and 93 μg/m3 respectively), through untargeted metabolomics. All participants underwent 8 rounds of physical examination to assess cardiopulmonary function and collect plasma samples for metabolomic analysis. Air purifiers were installed in 40 dormitories of the 78 participants for 14 days. Plasma metabolomics identified 25 differential metabolites associated with exposure to PM2.5 and none associated with exposure to PM10. Of the 78 students, 9 were considered susceptible to exposure to PM2.5 because they had diastolic pressure (DP) and forced levels vital capacity (CVF) significantly associated with the variation of PM2.5. By comparing their plasma metabolic profile to that of healthy individuals, 6 differential metabolites were identified: (lysoPC (P-20: 0), lysoPC (P-18: 1 (9z)), lysoPC (20: 1), lysoPC ( O -16:0), choline and found 1,3-diphenylprop-2-en-1-one). These findings pointed out that long-term exposure to PM2.5 leads to changes in phospholipid catabolism. Furthermore, LysoPC (P-20: 0) and LysoPC (P-18: 1 (9z)) increased significantly after the air purification intervention. Thus, the authors suggested the use of the six discriminating metabolites as potential biomarkers to identify individuals sensitive to exposure to PM 2.5; and LysoPC (P-20:0) and LysoPC (P-18:1 (9z)); as biomarkers of exposure to PM2.5.
The herein described studies highlight the importance of metabolomics as tool to assess the impact of environmental toxin exposure. Thus, this research approach is expected to replace epidemiological questionnaires that oftentimes underestimate environmental factors assessed by epidemiological studies.
Conclusions
The herein literature review sought to present the following data: important concepts of metabolome analysis; the steps of both untargeted and targeted metabolomics workflow; relevant publications on chemical exposure. It is worth noticing that metabolomics is a holistic method and, as such, it can provide early diagnosis of alterations in xenobiotic metabolism, which may contain metabolic patterns or signatures able to detect toxin exposure and to quantify environmental toxins, or to reveal patients’ health or disease status.
The present study compilation has shown that most studies are based on metabolome analysis through mixed methods, mainly LC-MS and GC-MS. Advances in analytical methods and the recent improvement in data processing tools have contributed to the evolution of metabolomics as scientific field. Moreover, these factors have helped integrating metabolomics to studies on the relationship among toxic mechanisms, exposure to environmental toxins and early discovery of biomarkers. Yet, the chemical exposure field — mainly chronic low-level exposure to chemical agents — remains largely unexplored. Therefore, omics-based biomarkers should be urgently applied to clinical decision-making, mainly to help protecting, diagnosing and recovering human health.
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Availability of Data and Materials
Not applicable.
Funding
Not applicable.
Authors' Contributions
MPRM was responsible for the bibliographical survey and writing of the original draft. LCA carried out the final writing and editing.
References
- 1.Prüss-Ustün A, Wolf J, C, Bos R, Neira M. (2016) Organization, W. H.Preventing Disease through Healthy Environments: A Global Assessment of the Environmental Burden of Disease. 259.
- 2.M T Smith, R De, S I Daniels. (2015) Using Exposomics to Assess Cumulative Risks and Promote Health.Environ. , Mol 56(9), 10-1002.
- 3.C P Wild. (2005) Complementing the Genome with an ‘“ Exposome ”’: The Outstanding Challenge of Environmental Exposure Measurement in Molecular Epidemiology. , Cancer Epidemiol Biomarkers Prev 14, 1847-1851.
- 4.S M Rappaport, Martyn T, S. (2012) . Environment and Disease Risks2010 https://doi.org/10.1126/science.1192603 .
- 6.Vineis P, Chadeau-hyam M, Gmuender H, Gulliver J, Herceg Z et al. (2017) . International Journal of Hygiene and The Exposome in Practice Design of the EXPOsOMICS Project.220 142-151.
- 7.A R Dunn, Connell K M S O, C. (2019) Neuroscience and Biobehavioral Reviews Gene-by-Environment Interactions. in Alzheimer ’ s Disease and Parkinson ’ s Disease.Neurosci. Biobehav. Rev.103: 7380.
- 8.Hemminki K, Bermejo Lorenzo, Försti J, A. (2006) The Balance between Heritable and Environmental Aetiology of Human Disease.Nat. , Rev 7(12), 958-965.
- 9.M, D I Walker, Vermeulen R, Chadeau-hyam M, D P Jones et al. (2019) . The Exposome Molecules to Populations.Annu. Rev. ofPharmacology Toxicol.59 107-127.
- 10. (2019) Organização Mundial da Saúde. Dez Ameaças à Saúde Que a OMS Combaterá Em2019. Ten health threats that WHO will fight in2019.
- 11.C P Wild. (2011) Future Research Perspectives on Environment and Health The Requirement for a More Expansive Concept of Translational Cancer Research.Environ. , Heal.10(Suppl 1, 1-4.
- 12.Aardema M, Grego J T Mac. (2003) Toxicology and Genetic Toxicology in the New Era of " Toxicogenomics ": Impact of " -Omics ". Technologies.499 13-25.
- 13.M T Smith, Vermeulen R, Li G, Zhang L, Lan Q et al. (2005) . Use of ‘ Omic ’ Technologies to Study Humans Exposed to Benzene.154 123-127.
- 14.Buesen R, B N Chorley, da Silva Lima, Daston B, Deferme G et al. (2017) Applying ’omics Technologies in Chemicals Risk Assessment: Report of an ECETOC Workshop. InRegulatory Toxicology and Pharmacology;. 91, 3-13.
- 15. (2008) European Centre for Ecotoxicology and Toxicology of Chemicals. Workshop on the Application of ‘Omic Technologies. in Toxicology and Ecotoxicology: Case Studies and Risk Assessment. InThe Application of ’omic Technologies in Toxicology and Ecotoxicology; Belgium 1-70.
- 16.Vineis P, A E Khan, Vlaanderen J, Vermeulen R. (2009) The Impact of New Research Technologies on Our Understanding of Environmental Causes of Disease The Concept of Clinical Vulnerability.Environ. 8(54), 10-1186.
- 17.Canuto G A B, J L Costa, Cruz P L R, S A Rosa, E D Policy et al. (2018) . METABOLÔMICA: DEFINIÇÕES, ESTADO-DA-ARTE E APLICAÇÕES REPRESENTATIVAS.Quim. Nova41 75-91.
- 18.Chen S, Zhang M, Bo L, Li S, Hu L et al. (2018) . Metabolomic Analysis of the Toxic Effect of Chronic Exposure of Cadmium on Rat Urine.Environ. Sci. Pollut. Res.25 3765-3774.
- 19.Zhang J, Shen H, Xu W, Xia Y, D B Barr et al. (2014) Urinary Metabolomics Revealed Arsenic Internal Dose-Related Metabolic Alterations: A Proof-of-Concept Study in a Chinese Male Cohort.Environ. Sci. Technol. be48 12265-12274.
- 20.Gao P, Hou L, N D Denslow, Xiang P, L Q Ma. (2018) Human Exposure to Polycyclic Aromatic Hydrocarbons Metabolomics Perspective.Environ. Int.119(July) 466-477.
- 21.E L Schymanski, N C Baker, A J Williams, R, Trezzi J et al. (2019) . Environmental Science Neurodegeneration Using Cheminformatics and Challenges.Environ. Sci. Process. Impacts View21(9) 1426-1445.
- 22.Surowiec I, Karimpour M, Gouveia-figueira S, Wu J, Unosson J et al. (2016) Multi-Platform Metabolomics Assays for Human Lung Lavage Fluids in an Air Pollution Exposure Study.Anal. , Bioanal. Chem.4751–4764 10-1007.
- 23.J, J I Beier, K C Falkner, Wheeler B, C J Mcclain et al. (2016) Occupational Exposures at a Polyvinyl Chloride Production Facility Are Associ- Ated with Significant Changes to the Plasma Metabolome.Toxicol. , Appl. Pharmacol.https://doi.org/10.1016/j.taap.(2016).10.001
- 24.Suvagandha D, Nishijo M, Swaddiwudhipong W, Honda R. (2014) A Biomarker Found in Cadmium Exposed Residents of Thailand by Metabolome Analysis.Int. , J. Environ. Res. Public Health11 3661-3677.
- 25.Tzoulaki I, Ebbels T M D, Valdes A, Elliott P, J P Ioannidis. (2014) . A. Practice of Epidemiology Design and Analysis of Metabolomics Studies in Epidemiologic Research A Primer on -Omic Technologies.Am J Epidemiol180(2) 129-139.
- 26.J K Nicholson, Holmes E, J M Kinross, Ara W, Takats Z et al. (2012) . Metabolic Phenotyping in Clinical and Surgical Environments.Nature491 384-392.
- 27.Zhou J, Yin Y. (2016) Strategies for Large-Scale Targeted Metabolomics Quantification by Liquid Chromatography-Mass Spectrometry.Analyst141. 6362-6373.
- 28.D S Wishart, Y D Feunang, Marcu A, A C Guo, Liang K et al. (2018) HMDB 4.0 The Human Metabolome Database for2018.Nucleic Acids Res.46:. 608-617.
- 29.S H, W E Kraus, C B Newgard. (2012) Basic Science for Clinicians Metabolomic Profiling for the Identification of Novel Biomarkers and Mechanisms Related to Common Form and Function.Circulation126(9). 1110-1120.
- 30.J K Nicholson, J C Lindon, Holmes E. (1999) Metabonomics”: Understanding the Metabolic Responses of Living Systems to Pathophysiological Stimuli via Multivariate Statistical Analysis of Biological NMR Spectroscopic Data. , Xenobiotica 29(11), 1181-1189.
- 31.Fiehn O. (2001) Combining Genomics , Metabolome Analysis , and Biochemical Modelling to Understand Metabolic Networks.Comp Funct. Genom2 155-168.
- 32.A K Kosmides, Kamisoglu K, S E Calvano, S A Corbett, I P Androulakis. (2013) Metabolomic Fingerprinting: Challenges and Opportunities.Crit. , Rev. Biomed 41(3), 205-221.
- 33.Holmes E, R L Loo, Cloarec O, Coen M, Tang H et al. (2007) . , Detection of Urinary Drug Metabolite ( Xenometabolome ) Signatures in Molecular Epidemiology Studies via Statistical Total Correlation ( NMR ) Spectroscopy. Anal 79(7), 2629-2640.
- 34.Fabian E, Bordag N, Herold M, Kamp H, Krennrich G et al. (2016) . Metabolite pro Fi Les of Rats in Repeated Dose Toxicological Studies after Oral and Inhalative Exposure.Toxicol. Lett.255 11-23.
- 35.M A García-sevillano, García-barrera T, Navarro F, Abril N, Pueyo C. (2015) . Combination of Direct Infusion Mass Spectrometry and Gas Chromatography Mass Spectrometry for Toxicometabolomic Study of Red Blood Cells and Serum of Mice Mus Musculus after Mercury Exposure.J. Chromatogr. B985 75-84.
- 36.Hou Y, Cao C, Bao W, Yang S, Shi H et al. (2015) The Plasma Metabolic Profiling of Chronic Acephate Exposure in Rats via an Ultra-Performance Liquid Chromatography-Mass Spectrometry Based.Mol. Biosyst.11, 506–515. https://doi.org/10.1039/C4MB00523F
- 37.Li H, Wang M, Liang Q, Jin S, Sun X et al. (2016) Urinary Metabolomics Revealed Arsenic Exposure Related to Metabolic Alterations in General Chinese Pregnant Women.J. Chromatogr. Ahttps://doi.org/10.1016/j.chroma2016.12.007
- 38.Boyles M S P, Ranninger C, Reischl R, Rurik M, Tessadri R et al. (2016) Copper Oxide Nanoparticle Toxicity Profiling Using Untargeted Metabolomics.Part. Fibre Toxicol.13(1). 1-20.
- 39.Dong X, Zhang Y, Dong J, Zhao Y, Guo J et al. (2017) Urinary Metabolomic Profiling in Rats Exposed to Dietary Di. , ( 2-Ethylhexyl ) Phthalate ( DEHP ) Using Ultra-Performance Liquid Chromatography Quadrupole Time-of-Flight Tandem Mass Spectrometry ( UPLC / Q-TOF-MS ).Sci. Pollut 24(20), 16659-16672.
- 40.Khamis M M, D J Adamko, El-aneed A. (2014) . , MASS SPECTROMETRIC BASED APPROACHES IN URINE METABOLOMICS AND BIOMARKER DISCOVERY.Mass Spectrom 36(2), 1-20.
- 42.Pasikanti K K, P C Ho, Chan E C Y. (2008) . Gas Chromatography / Mass Spectrometry in Metabolic Profiling of Biological Fluids.J. Chromatogr. B871 202-211.
- 43.M V Lind, O I Savolainen, A B Ross. (2016) The Use of Mass Spectrometry for Analysing Metabolite Biomarkers in Epidemiology Methodological and Statistical Considerations for Application to Large Numbers of Biological Samples. , Eur. J. Epidemiol
- 44.Forcisi S, Moritz F. (2013) . Liquid Chromatography – Mass Spectrometry in Metabolomics Research Mass Analyzers in Ultra High Pressure Liquid Chromatography Coupling.J. Chromatogr. A1292 51-65.
- 45.P S Gromski, Muhamadali H, D I Ellis, Xu Y, Correa E et al. (2015) Analytica Chimica Acta A Tutorial Review Metabolomics and Partial Least Squares-Discriminant Analysis – a Marriage of Convenience or a Shotgun Wedding.Anal. , Chim. Acta879 10-23.
- 46.Uppal K, D I Walker, Liu K, Li S, Go Y et al. (2016) . Computational Metabolomics A Framework for the Million Metabolome.Chem Res Toxicol29(12) 1956-1975.
- 47.Griffin J, Bollard M. (2004) Metabonomics: Its Potential as a Tool in Toxicology for Safety Assessment and Data Integration.Curr. , Drug 5(5), 10-2174.
- 48.Dudzik D, Barbas-bernardos C, García A, Barbas C. (2018) Quality Assurance Procedures for Mass Spectrometry Untargeted Metabolomics . a Review.J. , Pharm. Biomed. Anal.147 149-173.
- 49.Chaleckis R, Murakami I, Takada J, Kondoh H, Yanagida M. (2016) . , Individual Variability in Human Blood Metabolites Identifies Age-Related Differences.Proc NatlAcad Sci U S 113(16), 4252-4259.
- 50.E A Thévenot, Roux A, Xu Y, Ezan E, Junot C. (2015) Analysis of the Human Adult Urinary Metabolome Variations with Age. Body Mass Index, and Gender by Implementing a Comprehensive Work Fl Ow for Univariate and OPLS Statistical Analyses.J. Proteome Res14(8) 3322-3335.
- 51.M C Walsh, G A Mcloughlin, H M Roche, J F Ferguson, C A Drevon et al. (2014) . Impact of Geographical Region on Urinary Metabolomic and Plasma Fatty Acid Profiles in Subjects with the Metabolic Syndrome across Europe The , LIPGENE Study British Journal of Nutrition. Br J Nutr 111(3), 424-431.
- 52.C S Sasaqui, Lima K S C, R B Sousa, Araújo N A De, Pinto J C C S et al. (2018) Metabolomics of Physical Exercise: A Powerful Assessment Tool.Rev. , Virtual 10(5), 1207-1224.
- 53.Wurtz P, Wang Q, A J Kangas, R C, Skarp J et al. (2014) Metabolic Signatures of Adiposity in Young Adults Mendelian Randomization Analysis and Effects of Weight Change.PLoS Med11(12). e1001765. https://doi.org/10.1371/journal.pmed.1001765
- 54.Mastrangelo A, G Á Martos-moreno, García A, Barrios V, F J Rupérez et al. (2016) Insulin Resistance in Prepubertal Obese Children Correlates with Sex-Dependent Early Onset Metabolomic Alterations.Int. , J 40(10), 1494-1502.
- 55.Hertel J, Friedrich N, Wittfeld K, Pietzner M, Budde K et al. (2016) . Measuring Biological Age via Metabonomics – the Metabolic Age-Score Measuring Biological Age via Metabonomics – the Metabolic Age-Score.J Proteome Res.15(2) 400-410.
- 56.Scalabre A, Jobard E, Demède D, Gaillard S, Mouriquand P et al. (2017) . Evolution of Newborns’ Urinary Metabolomic Profiles According to Age and Growth Evolution of Newborns Urinary Metabolomic Profiles According to Age and Growth.J Proteome Res.16(10) 3732-3740.
- 57.C M Slupsky, K N Rankin, Wagner J, Fu H, Chang D et al. (2007) . Investigations of the Effects of Gender , Diurnal Variation , and Age in Human Urinary Metabolomic Profiles.Anal Chem79(18) 6995-7004.
- 58.Roux A, Lison D, Junot C, Heilier J. (2011) Applications of Liquid Chromatography Coupled to Mass Spectrometry-Based Metabolomics in Clinical Chemistry and Toxicology A Review.Clin. 44(1), 119-135.
- 59.M A Fernández-Peralbo, Castro M D L de. (2012) Preparation of Urine Samples Prior to Targeted or Untargeted Metabolomics Mass-Spectrometry Analysis.Trends Anal. Chem.41, 75–85. https://doi.org/10.1016/j.trac.(2012).08.011
- 60.Wu Y, Li L. (2016) . Sample Normalization Methods in Quantitative Metabolomics.J. Chromatogr. A1430 80-95.
- 61.D J Beale, F R Pinu, K A, M, V K Narayana et al. (2018) Review of Recent Developments in GC–MS Approaches to Metabolomics-Based Research.Metabolomics14(11). 1-31.
- 62.Hendriks M M W B, Eeuwijk F A Van, R H Jellema, J A Westerhuis, T H Reijmers et al. (2011) Data-Processing Strategies for Metabolomics Studies.Trends Anal. 30(10), 1685-1698.
- 63.Raterink R, P W Lindenburg, R J Vreeken, Ramautar R, Hankemeier T. (2014) . Recent Developments in Sample-Pretreatment Techniques for Mass Spectrometry-Based Metabolomics.Trends Anal. Chem.61 157-167.
- 64.Castro M D L de, F P Capote. (2017) . , THE ANALYTICAL PROCESS TO SEARCH FOR METABOLOMICS BIOMARKERS.J. Pharm. Biomed. Anal.https://doi.org/10.1016/j.jpba.2017.06.073
- 65.V L Stevens, Hoover E, Wang Y, K A Zanetti. (2019) Pre-Analytical Factors That Affect Metabolite Stability in Human Urine, Plasma, and Serum: A Review.Metabolites9(8). 156.
- 66.Lu W, Su X, M S Klein, I A Lewis, Fiehn O et al. (2017) Metabolite Measurement Pitfalls to Avoid and Practices to Follow.Annu. Rev. Biochem.86 277-304.
- 67.Pinto J, Domingues M R M, Galhano E, Pita C, Almeida M do C et al. (2014) . Human Plasma Stability during Handling and Storage: Impact on NMR Metabolomics.Analyst139(5) 1168-1177.
- 68.T Ž Semren, I B Karaconji, Safner T, Brajenovic N, B T Lovakovic et al. (2018) Gas Chromatographic-Mass Spectrometric Analysis of Urinary Volatile Organic Metabolites. Optimization of the HS-SPME Procedure and Sample Storage Conditions.Talanta176: 537-543.
- 69.Trezzi J, Jäger C, Galozzi S, Barkovits K, Marcus K et al. (2017) . MethodsX Metabolic pro Fi Ling of Body Fl Uids and Multivariate Data Analysis.4, 95–103. https://doi.org/10.1016/j.mex.(2017).02.004 .
- 70.Garcia A, Barbas C. (2011) Gas Chromatography-Mass Spectrometry (GC-MS)-Based. , Metabolomics.Metab 708(4), 10-1007.
- 71.B-J W Robertson, Kim Y-M, E M Zink, K A Hallaian, Zhang Q et al. (2015) . A Statistical Analysis of the Effects of Urease Pre-Treatment on the Measurement of the Urinary Metabolome by Gas Chromatography-Mass Spectrometry.Metabolomics10(5) 897, 10-1007.
- 72.Palmas F, Mussap M, Fattuoni C. (2018) Urine Metabolome Analysis by Gas Chromatography. Mass Spectrometry ( GC – MS ): Standardization and Optimization of Protocols for Urea Removal and Short-Term Sample Storage.Clin. Chim. Acta485(July) 236-242.
- 73.Orata F. (2012) Derivatization Reactions and Reagents for Gas Chromatography Analysis.Adv. Gas Chromatogr. , Prog. Agric. Biomed. Ind. Appl.83–108
- 74.Souza ARL. (2018) . , ABORDAGEM METABOLÔMICA NO ESTUDO DE EXPOSIÇÃO GESTACIONAL À FUMAÇA DE Cannabis Sativa EM COBAIAS, UNIVERSIDADE DE SÃO PAULO
- 75.Emwas A-H, Roy R, R T McKay, Tenori L, Saccenti E et al. (2019) NMR Spectroscopy for Metabolomics Research.Metabolites9(7). 123.
- 76.Ramautar R, G W Somsen, Jong G J de. (2017) . CE-MS for Metabolomics: Developments and Applications in the Period 40(1), 165-179.
- 77.Wang S, I A Blair, Mesaros C. (2019) . Analytical Methods for Mass Spectrometry-Based Metabolomics Studies.Adv. Exp. Med. Biol.1140 635-647.
- 78.F M Lanças. (2013) A Cromatografia Líquida Moderna e a Espectrometria de m Assas Finalmente “ Compatíveis ”? II . A Escolha Do Analisador de. 5(1), 27-46.
- 79.Han J, Datla R, Chan S. (2009) . Mass Spectrometry-Based Technologies for High-Throughput Metabolomics.Bionalaysis1(9) 1665-1684.
- 80.Habchi B, Alves S, Paris A, D N Rutledge, Rathahao-paris E. (2016) How to Really Perform High Throughput Metabolomic Analyses Efficiently Anal. , Chem.85 128, 10-1016.
- 81.F M Lanças. (2009) A Cromatografia Líquida Moderna e a Espectrometria de Massas Finalmente “. , Compatíveis 1(2), 35-61.
- 82.J M Halket, Waterman D, A M Przyborowska, Patel R K P, P D Fraser.. 2005,Chemical Derivatization and Mass Spectral Libraries in Metabolic Profiling by GC / MS and LC / MS / MS.J. Exp. Bot.56(410) 219-243.
- 83.Mastrangelo A, Ferrarini A, Rey-stolle F, García A, Barbas C. (2015) From Sample Treatment to Biomarker Discovery A Tutorial for Untargeted Metabolomics Based on GC-. , ( EI ) -Q-MS.Anal. Chim. Acta900 21-35.
- 84.Han T, Yang Y, Zhang H, K P Law. (2017) Analytical Challenges of Untargeted GC-MS-Based Metabolomics and the Critical Issues. in Selecting the Data Processing Strategy [ Version 1 Referees 2 Approved ] Referee Status No 0, 1-17.
- 85.Fiehn O. (2017) Metabolomics by Gas Chromatography-Mass Spectrometry: The Combination of Targeted and Untargeted Profiling.Curr Protoc Mol Biol.114.
- 86.B M Hounoum, Blasco H, Emond P, Mavel S. (2016) . Liquid Chromatography – High-Resolution Mass Spectrometry-Based Cell Metabolomics Experimental Design , Recommendations , and Applications.Trends Anal. Chem.75 118-128.
- 87.W B Dunn, D I Broadhurst, Edison A, Guillou C, M R Viant et al. (2017) Quality Assurance and Quality Control Processes Summary of a Metabolomics Community Questionnaire.Metabolomics.
- 88.Broadhurst D, Goodacre R, S N Reinke, Kuligowski J, I D Wilson et al. (2018) . Guidelines and Considerations for the Use of System Suitability and Quality Control Samples in Mass Spectrometry Assays Applied in Untargeted Clinical Metabolomic Studies.Metabolomics14(6) 1-17.
- 89.W B Dunn, Broadhurst D, Begley P, Zelena E, Francis-mcintyre S et al. (2011) Procedures for Large-Scale Metabolic Profiling of Serum and Plasma Using Gas Chromatography and Liquid Chromatography Coupled to Mass Spectrometry. https://doi.org/10.1038/nprot.(2011).335
- 90.Piovezan M. (2014) . , E CARACTERIZAÇÃO QUÍMICA DE EXTRATO DE AMORA PRETA (Rubus Fruticosus Var. Lochness) E SEU EFEITO SOBRE A DIETA HIPERLIPÍDICA EM RATOS MEDIANTE ANÁLISE METABOLÔMICA
- 91.Misra B B, J F Fahrmann, Grapov D. (2017) . Review of Emerging Metabolomic Tools and Resources 2015-2016. Electrophoresis 38(18), 2257-2274.
- 92.Kusonmano K, Vongsangnak W. (2016) Informatics for Metabolomics. InTranslational Biomedical Informatics. Advances in Experimental Medicine and Biology; , Ed.; Singapore 91-115.
- 93.Yi L, Dong N, Yun Y, Deng B, Ren D et al. (2016) . Chemometric Methods in Data Processing of Mass Spectrometry-Based Metabolomics: A Review.Anal. Chim. Acta914 17-34.
- 94.Pang Z, Chong J, Zhou G, Morais D A de L, Chang L et al. (2021) . MetaboAnalyst5.0: Narrowing the Gap between Raw Spectra and Functional Insights.NucleicAcids Res.1–9. https://doi.org/10.1093/nar/gkab382 .
- 95.Chong J, D S Wishart, Xia J. (2019) Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. , Curr. Protoc. Bioinforma 68(86), 1-128.
- 96.Du J, Yuan Z, Ma Z, Song J, Xie X et al. (2014) KEGG-PATH Kyoto Encyclopedia of Genes and Genomes-Based Pathway Analysis Using a Path.Mol. Biosyst.10. 2441-2447.
- 97.Caspi R, Billington R, C A Fulcher, I M Keseler, Kothari A et al. (2018) . The MetaCyc Database of Metabolic Pathways and Enzymes.NucleicAcidsRes.46: 633-639.
- 98.Kaever A, Landesfeind M, Feussner K. (2015) MarVis-Pathway Integrative and Exploratory Pathway Analysis of Non-Targeted Metabolomics Data.Metabolomics764–777.
- 99.Zhang B, Hu S, Baskin E, Patt A, J K Siddiqui. (2018) RaMP A Comprehensive Relational Database of Metabolomics Pathways for Pathway Enrichment Analysis of Genes and Metabolites.Metabolites8(16). 1-15.
- 100.Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-lopes C et al. (2019) . Pathway Enrichment Analysis and Visualization of Omics Data Using G Pro Fi Ler , GSEA , Cytoscape and EnrichmentMap.Nat.Protoc .
- 101.Vineis P. (2018) From John Snow to Omics: The Long Journey of Environmental Epidemiology. , Eur. J. Epidemiol 33(4), 355-363.
- 103.Q N Van, T D Veenstra, H J Issaq. (2011) Metabolic Profiling for the Detection of Bladder Cancer.Curr. , Urol 12(1), 34-40.
- 104.Souza J M O, Carneiro M F H, Carolina A, Paulelli C, Grotto D et al. (2015) . , ARSÊNIO E ARROZ: TOXICIDADE, METABOLISMO E SEGURANÇA ALIMENTAR.Quim 38(1), 118-127.
- 106.Li Y, Lu X, Yu N, Li A, Zhuang T et al. (2021) . Exposure to Legacy and Novel Perfluoroalkyl Substance Disturbs the Metabolic Homeostasis in Pregnant Women and Fetuses: A Metabolome-Wide Association Study.Environ. Int.156 106627-10.
- 107.Yang X, Zhang M, Lu T, Chen S, Sun X et al. (2020) . Metabolomics Study and Meta-Analysis on the Association between Maternal Pesticide Exposome and Birth Outcomes.Environ. Res.182: 109087.
- 108.Wang F, Zhang H, Geng N, Ren X, Zhang B et al. (2018) . A Metabolomics Strategy to Assess the Combined Toxicity of Polycyclic Aromatic Hydrocarbons ( PAHs ) and Short-Chain Chlorinated Paraf Fi Ns.Environ. Pollut.234 572-580.
- 109.Wang F, Zhang H, Geng N, Ren X, Zhang B et al. (2018) A Metabolomics Strategy to Assess the Combined Toxicity of Polycyclic Aromatic Hydrocarbons (PAHs) and Short-Chain Chlorinated Paraffins (SCCPs).Environ. Pollut.234, 572–580. https://doi.org/10.1016/j.envpol.(2017).11.073
- 110.D G Madruga, R M Ubeda, J M Terroba, Saúl G, Cambero J P G. (2019) Particle ‐ Associated Polycyclic Aromatic Hydrocarbons in a Representative Urban Location. ( Indoor ‐ Outdoor ) from South Europe Assessment of Potential Sources and Cancer Risk to Humans.Indoor Air29(5) 817-827.
- 111.Raffy G, Mercier F, Blanchard O, Derbez M, Dassonville C et al. (2016) . Semi-Volatile Organic Compounds in the Air and Dust of 30 French Schools A Pilot Study. 1–14. https://doi.org/10.1111/ina.12288 .
- 112.Wang Z, Zheng Y, Zhao B, Zhang Y, Liu Z et al. (2015) Human Metabolic Responses to Chronic Environmental Polycyclic Aromatic Hydrocarbon Exposure by a Metabolomic Approach.J. Proteome Res. 14(6), 2583-2593.
- 113. (2012) International Agency for Research on Cancer. Cadmium and Cadmium Compounds. (Group 2A).IARC Monogr. Int. Agency Res. Cancer.100 121-145.
- 114.Nishijo M, Nakagawa H, Suwazono Y, Nogawa K, Kido T. (2017) . Causes of Death in Patients with Itai-Itai Disease Suffering from Severe Chronic Cadmium Poisoning A Nested Case – Control Analysis of a Follow-up Study in Japan.BMJ Open7(7): 015694.
- 115.Nishijo M, Nakagawa H, Suwazono Y, Nogawa K, Sakurai M et al. (2018) . Cancer Mortality in Residents of the Cadmium-Polluted Jinzu River Basin in Toyama, Japan.Toxics6(2) 1-11.
- 116.Zeng T, Liang Y, Chen J, Cao G, Yang Z et al. (2021) Urinary Metabolic Characterization with Nephrotoxicity for Residents under Cadmium Exposure.Environ. Int.154 106646.
- 117.Haghani A, Johnson R, Safi N, Zhang H, Thorwald M et al. (2020) . Toxicity of Urban Air Pollution Particulate Matter in Developing and Adult Mouse Brain: Comparison of Total and Filter-Eluted Nanoparticles.Environ. Int.136: 105510.
- 118.Carmo M E G do, Kunizaki F C da C, Sousa N L da S, L. (2020) . Caracterização e Avaliação Da Toxicidade de MP10 Presentes Na Área Urbana de Catalão – GO Associados a Parâmetros Climatológicos.Rev. Process 14(27), 37-48.
- 119.R B Hayes, Lim C, Zhang Y, Cromar K, Shao Y et al. (2020) PM2.5 Air Pollution and Cause-Specific Cardiovascular Disease Mortality. , Int. J. Epidemiol 49(1), 25-35.
- 120.J D Newman, D L Bhatt, Rajagopalan S, J R Balmes, Brauer M et al. (2020) Cardiopulmonary Impact of Particulate Air Pollution in High-Risk Populations:. , JACC State-of-the-Art Review. J. Am. Coll. Cardiol 76(24), 2878-2894.
